Introduction
With modern wearable muscle near-infrared spectroscopy (mNIRS) devices it is growing ever easier to collect local muscle oxygenation data during dynamic activities. The real challenge comes with deciding how to clean, filter, process, and eventually interpret those data.
The mnirs package aims to provide standardised, reproducible methods for processing and analysing NIRS data, helping practitioners detect meaningful signals from noise and improve our confidence in interpreting information and applying that information to the clients we work with.
In this vignette we will demonstrate how to:
📂 Read data files exported from commercially available wearable NIRS devices, and import NIRS channels into a standard data frame format with metadata, ready for further processing.
📊 Plot and visualise data frames of class
"mnirs".🔍 Retrieve metadata stored with data frames of class
"mnirs"to avoid repetitively specifying which channels to process.🧹 Detect and replace local outliers, invalid values, and interpolate across missing data.
⏱️ Resample data to a higher or lower sample rate, to correct irregular sampling periods or match the frequency of other data sources.
📈️ Apply digital filtering to optimise signal-to-noise ratio for the responses observed in our data.
⚖️ Shift and rescale across multiple NIRS channels, to normalise signal dynamic range while preserving absolute or relative scaling between muscle sites and NIRS channels.
🔀 Demonstrate a complete data wrangling process using pipe-friendly functions.
Note
mnirsis currently. Functionality may change! Stay updated on development and follow releases at github.com/jemarnold/mnirs.
{mnirs} is designed to process NIRS data, but it can be used to read, clean, and process other time series datasets which require many of the same processing steps. Enjoy!
📂 Read data from file
We will read an example data file with two NIRS channels from an incremental ramp cycling assessment recorded with Moxy muscle oxygen monitor.
First, install and load the mnirs package and other required libraries.
mnirs can be installed with remotes::install_github("jemarnold/mnirs").
The first function to call will almost always be read_mnirs(). This is used to read data from .csv or .xls(x) files exported from common wearable NIRS devices. It will extract and return the data table and metadata for further processing and analysis.
See read_mnirs() for more details.
read_mnirs()
-
file_pathSpecify the location of the data file, including file extension. e.g.
"./my_file.xlsx"or"C:/myfolder/my_file.csv".
Example data files
A few example data files are included in the
mnirspackage. File paths can be accessed withexample_mnirs()
-
nirs_channelsAt least one NIRS channel name must be specified from the data table in the file. Multiple channel names can be entered as a vector.
-
time_channelA time or sample channel name from the data table can be specified. If left blank, the function will attempt to identify the time column automatically, however the best practice is to specify the time_channel column explicitly.
-
event_channelOptionally, A channel can be specified which indicates events or laps in the data table.
These channel names are used to detect the data table within the file, and must match exactly with text strings in the file on the same row. We can rename these channels when reading data by specifying a named character vector:
nirs_channels = c(new_name1 = "original_name1",
new_name2 = "original_name2")-
sample_rateThe sample rate of the exported data file (in Hz) can either be specified explicitly, or it will be estimated from
time_channel.
Automatic detection usually works well unless there is irregular sampling, or thetime_channelis a count of samples rather than a time value. For example, Oxysoft exports a sample number rather than time values. In this specific case, the function will recognise and read the correct sample rate from the file metadata. However, in most cases sample_rate should be defined explicitly if known. -
add_timestampIf the
time_channelis detected as date-time format (e.g.; hh:mm:ss), by default it will be converted to numeric time in seconds. If specified, this option will preserve a seperate timestamp column of those date-time values. -
zero_timeIf
time_channelvalues start at a non-zero value, this option will re-calculate time starting from zero. Iftime_channelwas converted from date-time format, it will always be re-calculated from zero regardless of this option. -
keep_allBy default, only the channels explicitly specified above will be returned in a data frame. This option will return all columns detected from the data table in the file. Blank/empty columns will be omitted and duplicated or invalid column names will be repaired. Columns should be checked to confirm correct naming if duplicates are present.
-
verboseThis and many other
mnirsfunctions may return warnings and informational messages which are useful for troubleshooting and data validation. This option can be used to silence those messages.mnirsmessages can be silenced globally for a session by settingoptions(mnirs.verbose = FALSE).
## {mnirs} includes sample files from a few NIRS devices
example_mnirs()
#> [1] "artinis_intervals.xlsx" "moxy_intervals.csv"
#> [3] "moxy_ramp.xlsx" "portamon-oxcap.xlsx"
#> [5] "train.red_intervals.csv" "vo2master.csv"
## partial matching will error if matches multiple
try(example_mnirs("moxy"))
#> Error in example_mnirs("moxy") : ✖ Multiple files match "moxy":
#> ℹ Matching files: "moxy_intervals.csv" and "moxy_ramp.xlsx"
## call an example mNIRS data file
file_path <- example_mnirs("moxy_ramp")
data_table <- read_mnirs(
file_path,
nirs_channels = c(
smo2_right = "SmO2 Live", ## identify and rename channels
smo2_left = "SmO2 Live(2)"
),
time_channel = c(time = "hh:mm:ss"), ## date-time format will be converted to numeric
event_channel = NULL, ## left blank, not currently used in analysis
sample_rate = NULL, ## if blank, sample_rate will be estimated from time_channel
add_timestamp = FALSE, ## omit the date-time timestamp column
zero_time = TRUE, ## recalculate time values from zero
keep_all = FALSE, ## return only the specified data channels
verbose = TRUE ## show warnings & messages
)
#> ! Estimated `sample_rate` = 2 Hz.
#> ℹ Define `sample_rate` explicitly to override.
#> Warning: ! Duplicate or irregular `time_channel` samples detected.
#> ℹ Investigate at `time` = 211.99 and 1184.
#> ℹ Re-sample with `mnirs::resample_mnirs()`.
## Note the above info message that sample_rate was estimated correctly at 2 Hz ☝
## ignore the warnings about irregular sampling for now, we will resample later
data_table
#> # A tibble: 2,203 × 3
#> time smo2_right smo2_left
#> <dbl> <dbl> <dbl>
#> 1 0 54 68
#> 2 0.400 54 68
#> 3 0.960 54 68
#> 4 1.51 54 66
#> 5 2.06 54 66
#> 6 2.61 54 66
#> 7 3.16 54 66
#> 8 3.71 57 67
#> 9 4.26 57 67
#> 10 4.81 57 67
#> # ℹ 2,193 more rows📊 Plot mnirs data
mnirs data can be easily viewed by calling plot(). This generic plot function uses ggplot2 and will work on data frames generated or read by mnirs functions where the metadata contain class = *"mnirs"*.
## note the hidden plot option to display time values as `h:mm:ss`
plot(data_table, label_time = TRUE)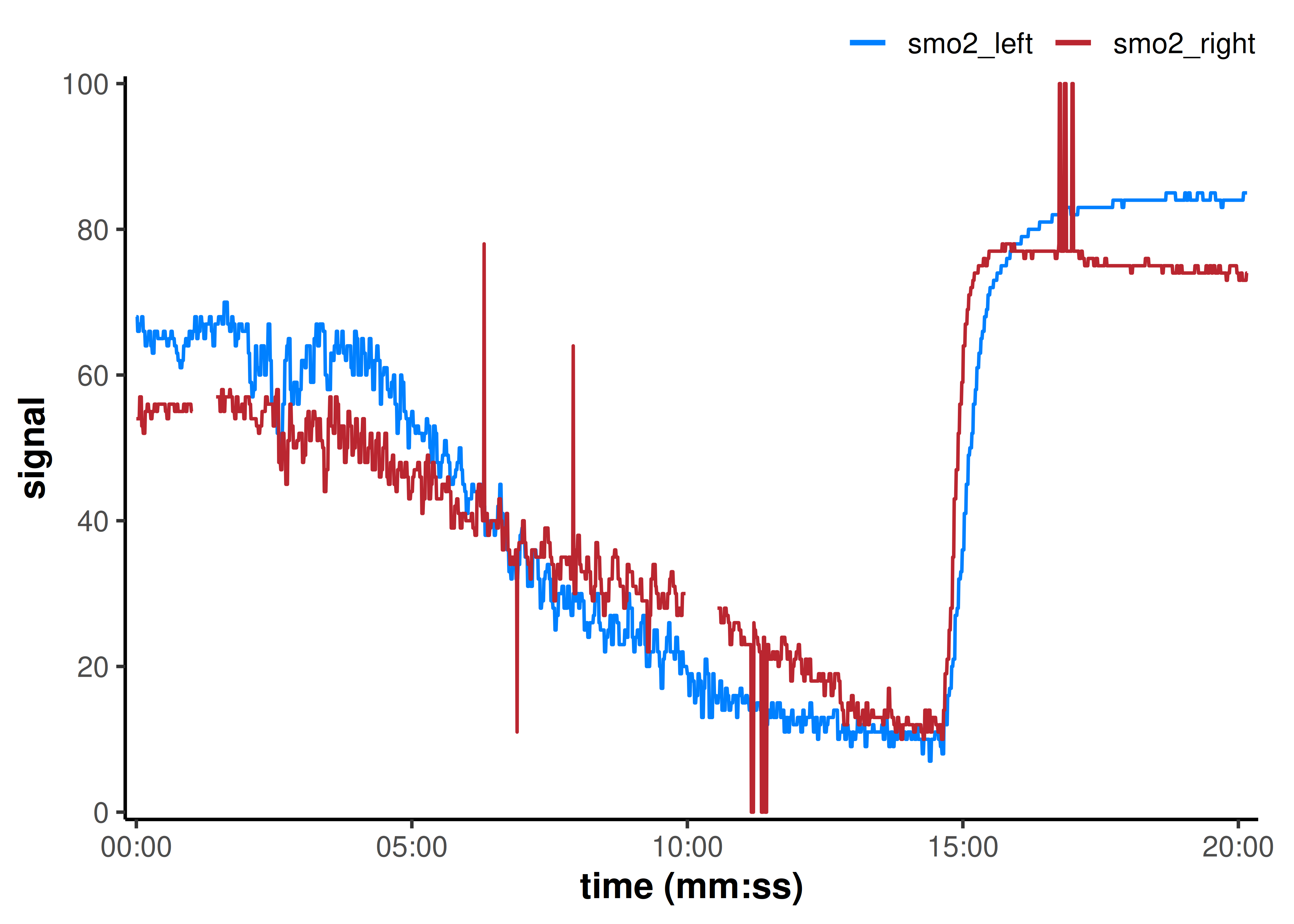
mnirs includes a custom *ggplot2* theme and colour palette available with theme_mnirs() and palette_mnirs(). See those documentation references for more details.
🔍 Metadata stored in mnirs data frames
Data frames generated or read by mnirs functions will return class = *"mnirs"* and contain metadata, which can be retrieved with attributes(data).
Instead of re-defining values like our channel names or sample rate, certain mnirs functions can automatically retrieve them from metadata. They can always be overwritten manually in subsequent functions, or by using a helper function create_mnirs_data().
See create_mnirs_data() for more details about metadata.
## view metadata (omitting item two, a list of row numbers)
attributes(data_table)[-2]
#> $class
#> [1] "mnirs" "tbl_df" "tbl" "data.frame"
#>
#> $names
#> [1] "time" "smo2_right" "smo2_left"
#>
#> $nirs_device
#> [1] "Moxy"
#>
#> $nirs_channels
#> [1] "smo2_right" "smo2_left"
#>
#> $time_channel
#> [1] "time"
#>
#> $sample_rate
#> [1] 2🧹 Replace local outliers, invalid values, and missing values
We can see some data issues in the plot above, so let’s clean those with a single wrangling function replace_mnirs(), to prepare our data for digital filtering and smoothing.
mnirs tries to include basic functions which work on vector data, and convenience wrappers which combine functionality and can be used on multiple channels in a data frame at once. Let’s explain the functionality of this data-wide function, and for more details about the vector-specific functions see replace_mnirs().
replace_mnirs()
-
dataData-wide functions take in a data frame, apply processing to all channels specified, then return the processed data frame.
mnirsmetadata will be passed to and from this function.mnirsfunctions are also pipe-friendly for Base R 4.1+ (|>) or magrittr (%>%) pipes to chain operations together (see below). -
nirs_channelsSpecify which column names in
datawill be processed, i.e. the response variables. If not specified, these channels will be retrieved frommnirsmetadata. Channels in the data but not explicitly specified will be passed through unprocessed to the returned data frame. -
time_channelsThe time channel can be specified, i.e. the predictor variable, or retrieved from
mnirsmetadata. -
invalid_values,invalid_above, orinvalid_belowSpecific invalid values can be replaced, e.g. if a NIRS device exports
0,100, or some other fixed value when signal recording is in error. If spikes or drops are present in the data, these can be replaced by specifying values above or below which to consider invalid. If left asNULL, no values will be replaced. -
outlier_cutoffLocal outliers can be detected using a cutoff calculated from the local median value. A default value of
3is recommended (i.e., ± 3 SD about the local median). If left asNULL, no outliers will be replaced. -
widthorspanLocal outlier detection and median interpolation use a rolling local window specified by one of either
widthorspan.widthdefines a number of samples centred around the local index being evaluated (idx), whereasspandefines a range of time in units oftime_channel. -
methodMissing data (
NA), invalid values, and local outliers specified above can be replaced via interpolation or fill methods; either"linear"interpolation, fill with local"median", or"locf"(“last observation carried forward”).NAs can be passed through to the returned data frame withmethod = "none". However, subsequent processing & analysis steps may return errors whenNAs are present. Therefore, it is good practice to deal with missing data early during data processing. -
verboseAs above, an option to toggle warnings and info messages.
data_cleaned <- replace_mnirs(
data_table, ## channels will be retrieved from metadata
invalid_values = 0, ## known invalid values in the data
invalid_above = 90, ## remove data spikes
outlier_cutoff = 3, ## recommended default value
width = 10, ## local window to detect local outliers and replace missing values
method = "linear", ## linear interpolation over `NA`s
verbose = TRUE
)
plot(data_cleaned, label_time = TRUE)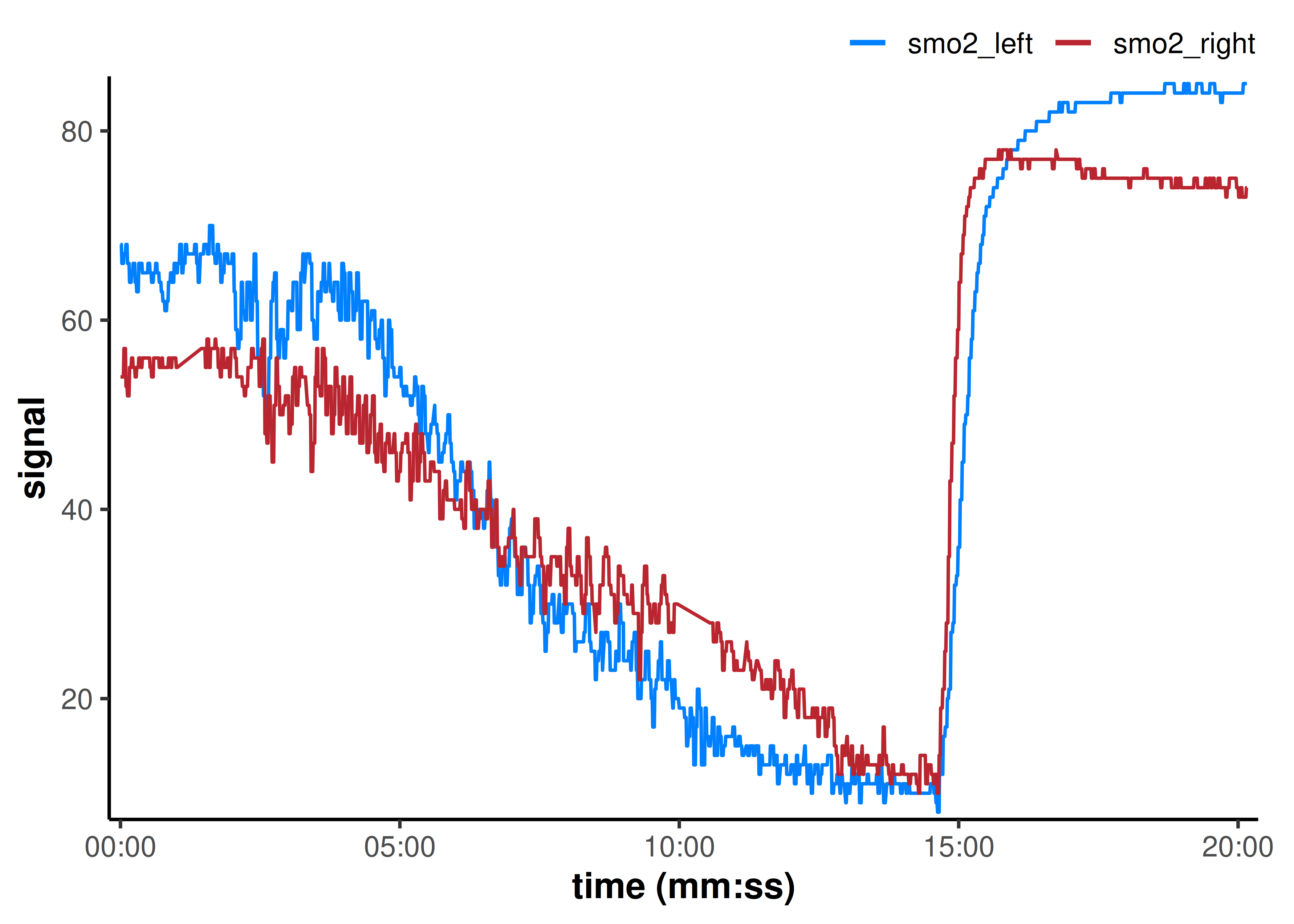
That cleaned up all the obvious data issues.
⏱️ Resample data
Say we have NIRS data recorded at 25 Hz, but we are only interested in exercise responses over a time span of 5-minutes, and our other outcome measure heart rate data are only recorded at 1 Hz anyway. It may be easier and faster to work with our NIRS data down-sampled from 25 to 1 Hz.
Alternatively, if we have something like high-frequency EMG data, we may want to up-sample our NIRS data to match samples for easier synchronisation and analysis (although, we should be cautious with up-sampling as this can artificially inflate statistical confidence with subsequent analysis or modelling methods).
resample_mnirs()
-
dataThis function takes in a data frame, applies processing to all channels specified, then returns the processed data frame.
mnirsmetadata will be passed to and from this function. -
time_channel&sample_rateIf the data contain
mnirsmetadata, these channels will be detected automatically. Or they can be specified explicitly. -
resample_rateResampling is specified as the desired number of samples per second (Hz). The default
resample_ratewill resample back to the existingsample_rateof the data. This can be useful to accomodate for irregular sampling with unequal time values. Linear interpolation is used to resampletime_channelto round values of thesample_rate.
data_resampled <- resample_mnirs(
data_cleaned, ## channels retrieved from metadata
resample_rate = 2, ## or leave blank as default `resample_rate = sample_rate`
method = "linear", ## linear interpolation across any new samples
verbose = TRUE ## will confirm the output sample rate
)
#> ℹ Output is resampled at 2 Hz.
## note the altered "time" values from the original data frame 👇
data_resampled
#> # A tibble: 2,419 × 3
#> time smo2_right smo2_left
#> <dbl> <dbl> <dbl>
#> 1 0 54 68
#> 2 0.5 54 68
#> 3 1 54 67.9
#> 4 1.5 54 66.0
#> 5 2 54 66
#> 6 2.5 54 66
#> 7 3 54 66
#> 8 3.5 55.9 66.6
#> 9 4 57 67
#> 10 4.5 57 67
#> # ℹ 2,409 more rowsUse
resample_mnirs()to smooth over irregular or skipped samplesIf we see a warning from
read_mnirs()about duplicated or irregular samples like we saw above, we can useresample_mnirs()to restoretime_channelto a regular sample rate and interpolate across skipped samples.
📈 Digital filtering
To improve the signal-to-noise ratio in our dataset without losing information, we should apply digital filtering to smooth the data.
Choosing a digital filter
There are a few digital filtering methods available in mnirs. Which option is best for you will depend in large part on the sample rate of the data and the frequency of the response/phenomena being observed.
Choosing filter parameters is an important processing step to improve signal-to-noise ratio and enhance our subsequent interpretations. Over-filtering the data can introduce data artefacts which can negatively influence the signal analysis and interpretations just as much as trying to analyse overly-noisy raw data.
It is perfectly valid to choose a digital filter by iteratively testing filter parameters until the signal or response of interest appears to be visually optimised with minimal data artefacts, to your satisfaction.
We will discuss the process of choosing a digital filter more in depth in another vignette <currently under development>.
filter_mnirs()
-
dataThis function takes in a data frame, applies processing to all channels specified, then returns the processed data frame.
mnirsmetadata will be passed to and from this function. -
nirs_channels,time_channel, &sample_rateIf the data contain
mnirsmetadata, these channels will be detected automatically. Or they can be specified explicitly. -
na.rmThis important argument is left as
FALSEby default. This function will return an error if any missing data (NA) are detected in the response variables (nirs_channels). Settingna.rm = TRUEwill bypass theseNAs and preserve them in the returned data frame, but this must be opted into explicitly in this function and elsewhere.
Smoothing-spline
-
method = "smooth_spline"The default non-parametric cubic smoothing spline is often a good first filtering option when exploring the data, and works well over longer time spans. For more rapid and repeated responses, a smoothing-spline may not work as well.
-
sparThe smoothing parameter of the cubic spline will be determined automatically by default, or can be specified explicitly. See
stats::smooth.spline()
Butterworth digital filter
-
method = "butterworth"A Butterworth low-pass digital filter (specified by
type = "low") is probably the most common method used in mNIRS research (whether appropriately or not). For certain applications such as identifying a signal with a known frequency (e.g. cycling/running cadence or heart rate), a pass-band or a different filter type may be better suited. -
typeThe filter type is specified as either
c("low", "high", "stop", "pass"). -
orderThe filter order number, specifying the number of passes the filter performs over the data. The default
order = 2will often noticably improve the filter over a single pass, however higher orders above ~4can begin to introduce artefacts, particularly at sharp transition points. -
WorfcThe cutoff frequency can be specified either as
W; a fraction (between[0, 1]) of the Nyquist frequency, which is equal to half of thesample_rateof the data. Or asfc; the cutoff frequency in Hz, where this absolute frequency should still be between 0 Hz and the Nyquist frequency.
For filtering vector data and more details about Butterworth filter parameters, see filter_butter().
Moving average
-
method = "moving_average"The simplest smoothing method is a moving average filter applied over a specified number of samples or timespan. Commonly, this might be a 5- or 15-second centred moving average.
-
widthorspanMoving average filtering occurs within a rolling local window specified by one of either
widthorspan.widthdefines a number of samples centred around the local index being evaluated (idx), whereasspandefines a range of time in units oftime_channel.
For filtering vector data and more details about the moving average filter, see filter_moving_average()
Apply the filter
Let’s try a Butterworth low-pass filter, and we’ll specify some empirically chosen filter parameters for these data. See filter_mnirs() for further details on each of these filtering methods and their respective parameters.
data_filtered <- filter_mnirs(
data_resampled, ## channels retrieved from metadata
method = "butterworth", ## Butterworth digital filter is a common choice
type = "low", ## specify a low-pass filter
order = 2, ## filter order number
W = 0.02, ## filter fractional critical frequency
na.rm = TRUE ## explicitly preserve any NAs
)
## we will add the non-filtered data back to the plot to compare
plot(data_filtered, label_time = TRUE) +
geom_line(
data = data_cleaned,
aes(y = smo2_left, colour = "smo2_left"), alpha = 0.4
) +
geom_line(
data = data_cleaned,
aes(y = smo2_right, colour = "smo2_right"), alpha = 0.4
)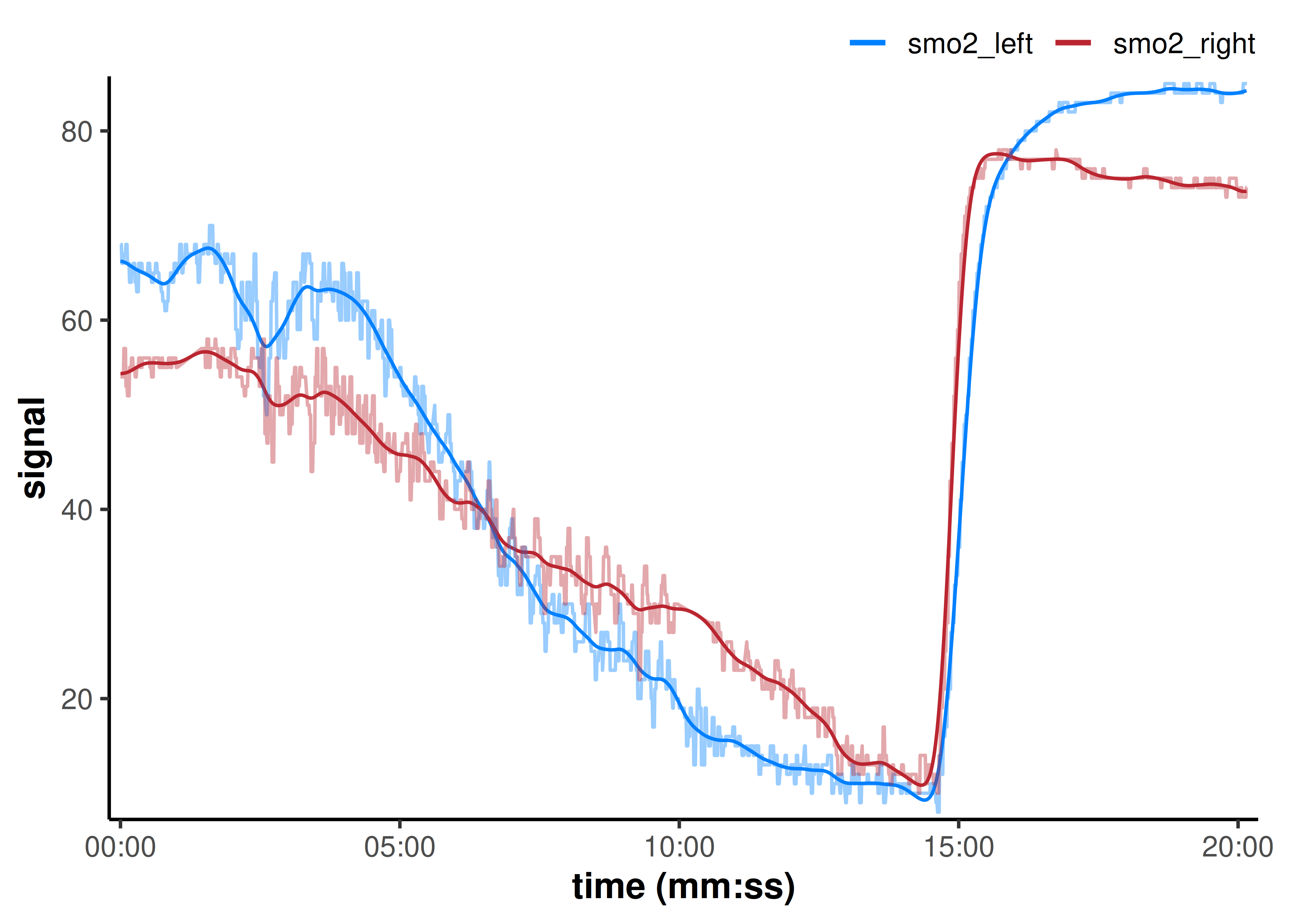
That did a nice job reducing the high-frequency noise in our data, while preserving the sharp edges, such as the rapid reoxygenation after maximal exercise.
⚖️ Shift and rescale data
NIRS values are not measured on an absolute scale (even, arguably percent (%) saturation). Therefore, we may need to adjust or calibrate our data to normalise NIRS signal values between muscle sites, individuals, trials, etc. depending on our intended comparison.
For example, we may want to set our mean baseline value to zero for all NIRS signals at the start of a recording. Or we may want to compare signal kinetics (the rate of change or time course of a response) after rescaling signal amplitudes to a common dynamic range.
These functions allow us to either shift NIRS values up or down while preserving the dynamic range (the absolute amplitude from minimum to maximum values) of our NIRS channels, or rescale the data to a new dynamic range with larger or smaller amplitude.
We can group NIRS channels together to preserve the absolute and relative scaling among certain channels, and modify that scaling between other groups of channels.
shift_mnirs()
-
dataThis function takes in a data frame, applies processing to all channels specified, then returns the processed data frame.
mnirsmetadata will be passed to and from this function. -
nirs_channelsIn these functions, channels should be grouped by providing a list (e.g.
list(c(A, B), c(C))) where each group will be shifted to a common scale, and separate scales between groups. The relative scaling between channels will be preserved within each group, but lost between groups.nirs_channelsshould be specified explicitly to ensure the intended grouping structure is returned. The defaultmnirsmetadata will group all NIRS channels together. -
time_channelIf the data contain
mnirsmetadata, this channels will be detected automatically. Or it can be specified explicitly -
toorbyThe shift amplitude can be specified by either shifting signals
toa new value, or shifting signalsbya fixed amplitude, given in units of the NIRS signals. -
widthorspanShifting can be performed on the mean value within a window specified by one of either
widthorspan.widthdefines a number of samples centred around the local index being evaluated (idx), whereasspandefines a range of time in units oftime_channel.
e.g.width = 1will shift from the single minimum sample, whereasspan = 1would shift from the minimum mean value of all samples within one second. -
positionSpecifies how we want to shift the data; either shifting the “min”, “max”, or “first” sample(s).
For this data set, we want to shift each NIRS channel so that the mean of the 2-minute baseline is equal to zero, which would then give us a change in deoxygenation from baseline during the incremental cycling protocol.
tidyverse-style channel name specification
This is a good time to note that here and in most
*mnirs*functions, data channels can be specified using tidyverse-style naming; Data frame column names can be specified either with quotes as a character string ("smo2"), or as a direct symbol (smo2).tidyselect support functions (e.g.
starts_with(),matches()) can also be used.
data_shifted <- shift_mnirs(
data_filtered, ## un-grouped nirs channels to shift separately
nirs_channels = list(smo2_left, smo2_right),
to = 0, ## NIRS values will be shifted to zero
span = 120, ## shift the first 120 sec of data to zero
position = "first"
)
plot(data_shifted, label_time = TRUE) +
geom_hline(yintercept = 0, linetype = "dotted")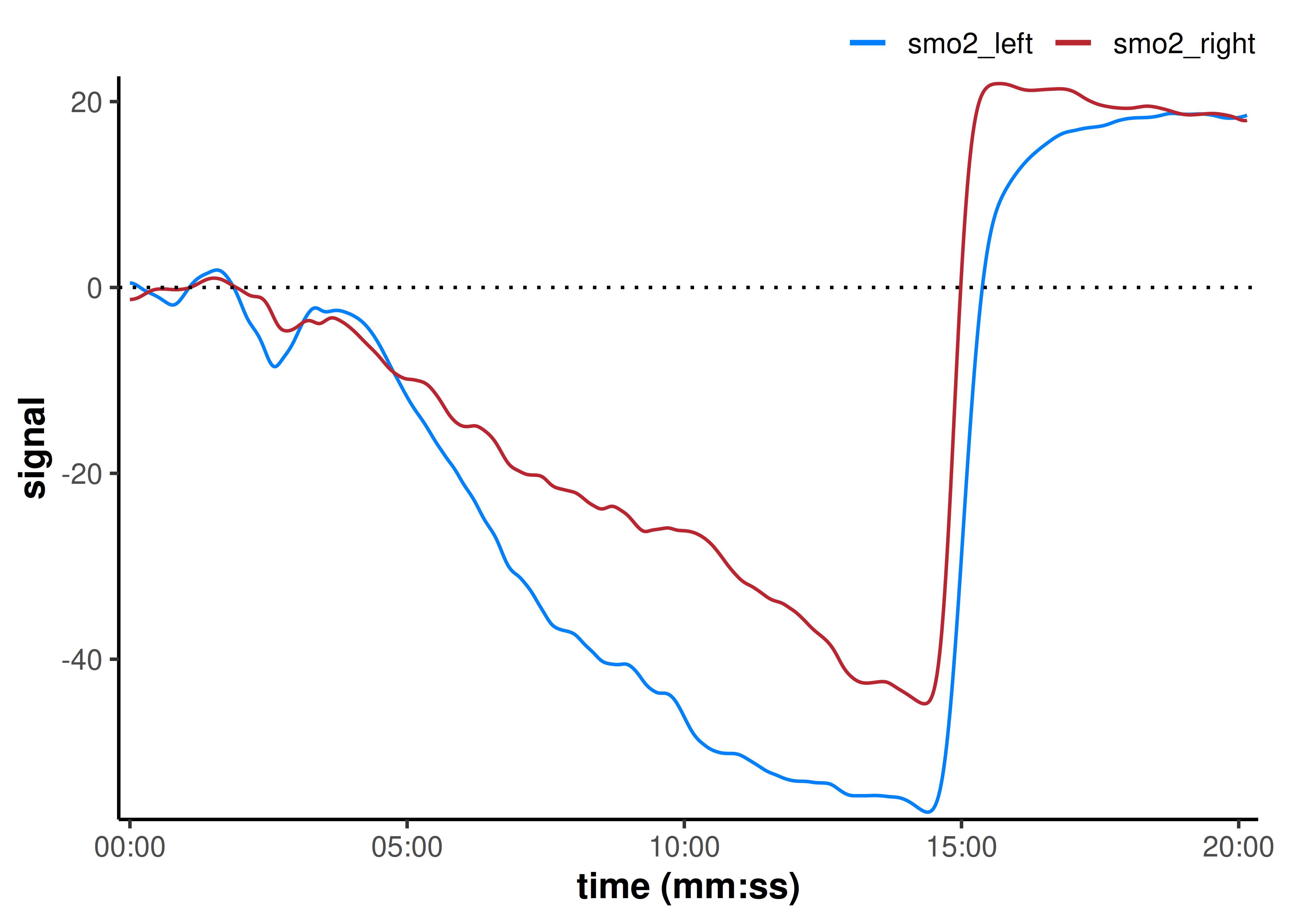
Before shifting, the minimum (end of exercise) values for smo2_left and smo2_right were similar, but the starting baseline values were different.
Whereas when we shift both baseline values to zero, we can see that the smo2_right signal has a smaller deoxygenation amplitude compared to the smo2_left signal.
We have to consider how our assumptions and processing decisions will influence our interpretations; by shifting both starting values, we are normalising for the starting condition of the two tissue sites and implicitly assuming that the baseline represents the same starting condition in both legs.
For example, this may be appropriate when we are more interested in the relative change (delta) in each leg during an intervention or exposure (often referred to as "∇SmO2").
rescale_mnirs()
We may also want to rescale our data to a new dynamic range, changing the units to a new amplitude.
-
dataThis function takes in a data frame, applies processing to all channels specified, then returns the processed data frame.
mnirsmetadata will be passed to and from this function. -
nirs_channelsChannels should be grouped by providing a list (e.g.
list(c(A, B), c(C))) where each group will be rescaled to a common range, and separate ranges between groups. The relative scaling between channels will be preserved within each group, but lost between groups.nirs_channelsshould be specified explicitly to ensure the intended grouping structure is returned. The defaultmnirsmetadata will group all NIRS channels together. -
rangeSpecifies the new dynamic range in the form
c(min, max). For example, if we want to calibrate each NIRS signal to their own observed ‘functional range’ during exercise, we could rescale them to 0-100%.
data_rescaled <- rescale_mnirs(
data_filtered, ## un-grouped nirs channels to rescale separately
nirs_channels = list(smo2_left, smo2_right),
range = c(0, 100) ## rescale to a 0-100% functional exercise range
)
plot(data_rescaled, label_time = TRUE) +
geom_hline(yintercept = c(0, 100), linetype = "dotted")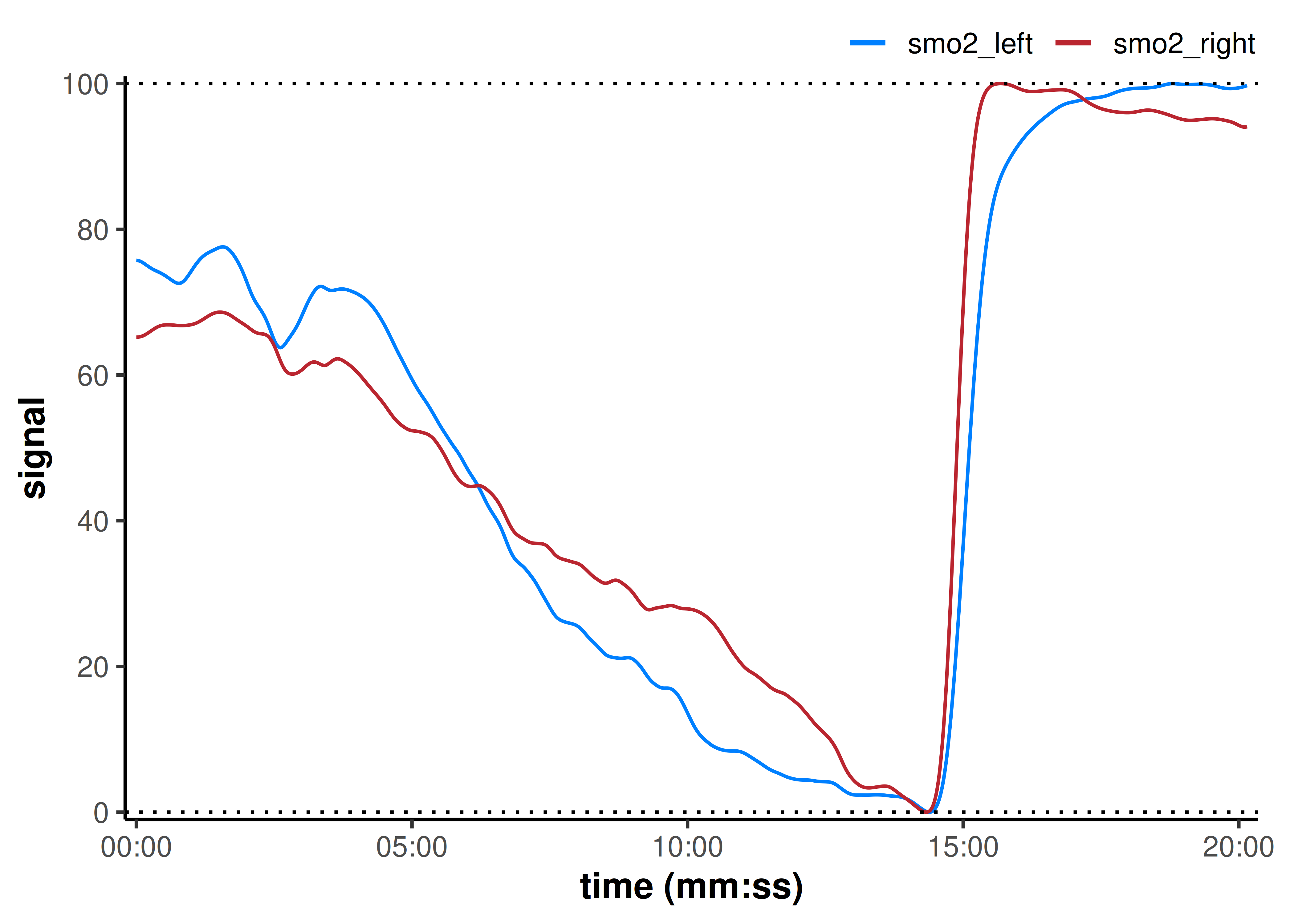
Here, our assumption is that during a maximal exercise task, the minimum and maximum values represent the functional capacity of each tissue volume being observed. So we rescale the functional dynamic range in each leg.
By normalising this way, we might lose meaningful differences captured by the different amplitudes between smo2_left and smo2_right, but we might be more interested in the trend or time course of each response.
Our interpretation may be that smo2_left appears to start at a slightly higher percent of it’s functional range, deoxygenates faster toward a minimum, and reaches a pseudo-plateau near maximal exercise. While smo2_right deoxygenates slightly slower and continues to deoxygenate until maximal task tolerance. Additionally, the right leg reoxygenates slightly faster than left during recovery. This might, for example, indicate exercise capacity differences between the limbs.
🔀 Pipe-friendly functions
Most mnirs functions can be piped together using Base R 4.1+ (|>) or magrittr (%>%). The entire pre-processing stage can easily be performed in a sequential pipe.
To demonstrate this, we’ll read a different example file recorded with Train.Red FYER muscle oxygen sensor and pipe it through each processing stage straight to a plot.
This is also a good time to demonstrate how to use the global mnirs.verbose argument to silence all warning & information messages, for example when dealing with a familiar dataset. We recommend leaving verbose = TRUE by default whenever reading and exploring a new file.
options(mnirs.verbose = FALSE)
read_mnirs(
example_mnirs("train.red"),
nirs_channels = c(
smo2_left = "SmO2 unfiltered",
smo2_right = "SmO2 unfiltered"
),
time_channel = c(time = "Timestamp (seconds passed)"),
zero_time = TRUE
) |>
resample_mnirs() |> ## default will resample to fix irregular samples
replace_mnirs(
invalid_above = 73,
outlier_cutoff = 3,
span = 7
) |>
filter_mnirs(
method = "butterworth",
order = 2,
W = 0.01,
na.rm = TRUE
) |>
shift_mnirs(
nirs_channels = list(smo2_left, smo2_right),
to = 0,
span = 60,
position = "first"
) |>
rescale_mnirs(
nirs_channels = list(c(smo2_left, smo2_right)), ## 👈 channels grouped together
range = c(0, 100)
) |>
plot(label_time = TRUE)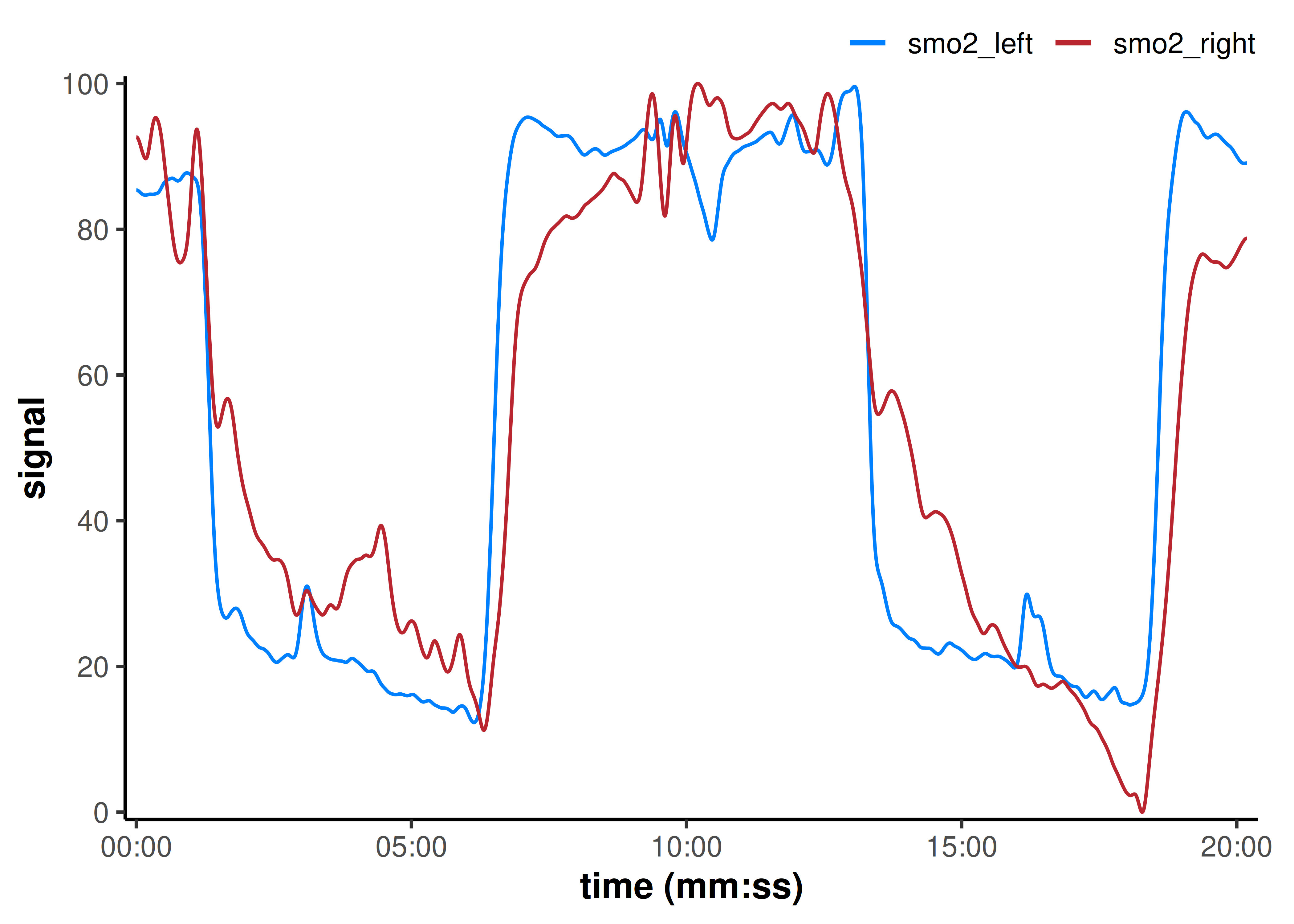
✅ Signal Processing
After the NIRS signal has been cleaned and filtered, it should be ready for further processing and analysis.
mnirs is being developed to include functionality for processing discrete intervals and events, e.g. reoxygenation kinetics, slope calculations for post-occlusion microvascular responsiveness, and critical oxygenation breakpoints.